Recent Articles
Categories
The Human Genome
The Human Genome Project (HGP) began in 1990 and remains one of the most major international biological endeavours of our time. Over the span of 13 years, researchers from 20 different centres across 6 countries, came together and successfully mapped nearly all 3 billion base pairs of the human genome with its approximately 30 000 genes [1]. Accompanying these advancements were the development of new DNA analysis technologies that could be used on massive genome-scale projects. The HGP has since fuelled the discovery of more than 1800 disease genes, and allowed for the development of now more than 2000 tests for various genetic conditions [2].
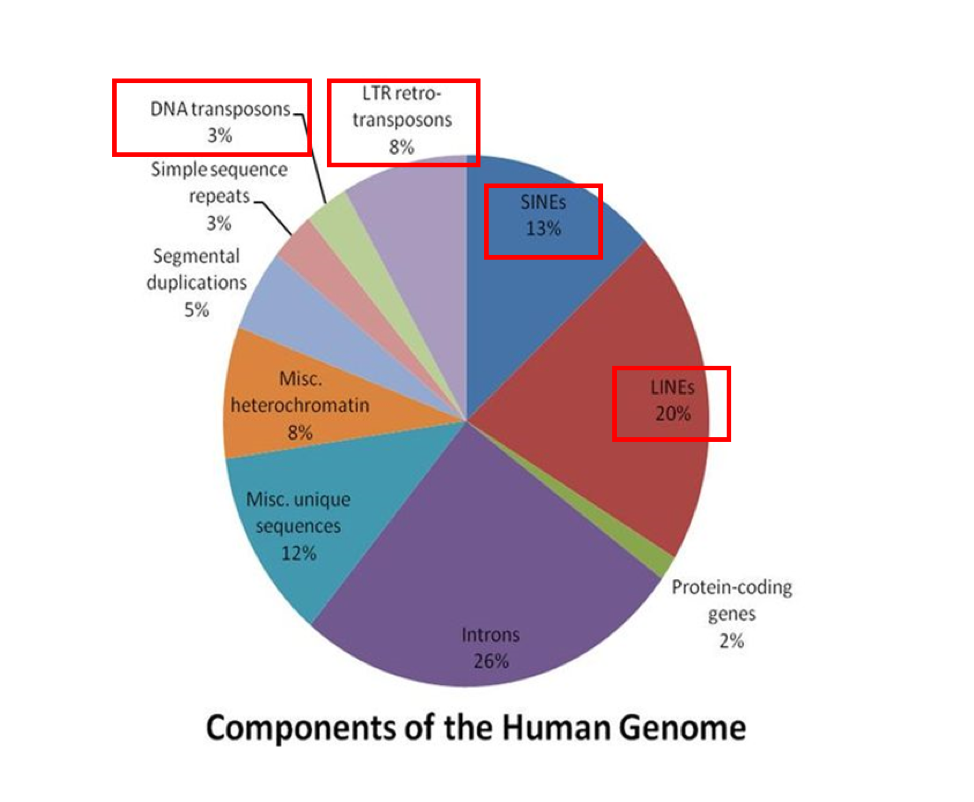
Figure 1. Proportions of various genomic components within the human genome with transposon portions boxed in red. Image retrieved and modified under a CC Attribution-Share Alike 3.0 Unported license.
Nonetheless, many of the intricacies of the human genome have yet to be fully unravelled. On average, two human beings will differ at about 1 in every 1000 DNA base pairs. Beneath all the genetic diversity of the human race, rests only 0.1% of our DNA sequence that makes each of us unique [3]. Our genetic identity is constantly changing and evolving and much of that variation has been the result of genomic rearrangements such as insertions, deletions, inversions, and duplications.
Copy-paste Genes
An interesting category of DNA elements that contributes immensely to genetic variation is what are known as transposable elements. Occupying close to half of the human genome, these elements can move around within the genome (Figure 1). A particular subclass of transposable elements called retrotransposons, use what is called the copy-paste mechanism for moving around the genome. Essentially, much like how you would copy and paste on a computer, these distinct DNA segments are able to replicate themselves and insert copies into new locations. The movement and regulation of retrotransposons is still largely a mystery, but retrotransposons are thought to play a role in everything from evolution to cancer.
Each and every one of our cells contains all the information necessary to create the incredibly complex organism that is the human being. Given the importance of the information it contains, it’s not surprising that we have developed safeguards against excessive movement of retrotransposons - one wrong move could lead to disaster. Many epigenetic defense mechanisms have been studied extensively; these mechanisms do not change the information stored in genes but instead regulate the expression of those genes. Some examples include modifying how tightly coiled portions of the DNA are, controlling access to the stored information; and DNA methylation, a process that essentially adds blockers to a strand of DNA to reduce how much it gets expressed.
One recently discovered epigenetic defense system involves small RNA-based elements called piRNAs. These piRNA complexes are made up of a single stranded piece of RNA bound to a special class of protein called piwi proteins - both encoded for naturally by the host genome. These RNA act as guides that direct the piRNA complex to specific complementary RNA transcripts, where the piwi-associated proteins can then catalyze their destruction [4].
Effects & Research of Transposons
Deregulation of transposable element activity can lead to many different problems for an organism. Dr. Denise Clark from UNB, is interested in the role transposable elements play in the creation of retrocopies which are essentially duplicates of genes that get re-inserted back into the genome with the mobile element acting as a carrier – these retrocopies can be active or inactive depending on where they insert. Dr. Clark uses fruit flies, or Drosophila, as a model organism to study genomic rearrangements because they have short life cycles, are easily grown and maintained, and produce a lot of offspring among many other reasons.
One particular system of hybrid dysgenesis in Drosophila has been of interest at the Clark lab, and involves the deregulation of I-type transposable elements that are thought to cause sterility in hybrid females. I-R hybrids are generated from what are called the inducer (I) and reactive (R) strains of Drosophila (Figure 2). Active I elements are present in the I strain but are thought to be mostly repressed by the host piRNA system. R strain individuals do not have I elements and therefore lack the piRNA defense mechanism. Sterility occurs in females that are the offspring of an I strain male is crossed with an R female. We call these SF females as opposed to RSF females which are the product of the reciprocal cross and who are fully fertile.
Figure 2. IR hybrids exhibit sterility in SF females generated from the dysgenic cross (left), whereas the reciprocal, non-dysgenic cross (right) produces fertile female offspring. Images of flies are by Madboy74 [CC0], retrieved from Wikimedia Commons.
We are essentially seeing two genetically identical females with different phenotypes, but the epigenetic action of the piRNA pathway provides us with a possible explanation. SF hybrid systems appear to have largely reduced maternal epigenetic protection from their R strain mothers leading to high levels of expression of paternally transmitted I elements. Genetically identical females, from the reverse R-I (male-female) crosses, known as RSF females, show much lower I element expression and are fully fertile emphasizing the maternally transmitted facet of piRNA protection [5].
Both SF and RSF hybrids lay a normal number of eggs which can be fertilized. Fertilized eggs follow a gonomeric type fertilization process, in which the paternal and maternal chromosomes enter mitosis as separate entities after pronuclear fusion. Studies have shown that in SF eggs fertilized by wild type sperm, only the maternal chromosomes have problems undergoing mitosis; these chromosomes either don’t connect properly before division or lag behind during the splitting process [6]. The understanding is that these meiotic defects are related to chromosome rearrangements and non-disjunctions generated from I element integration.
Some other intriguing features of the IR system involve age and temperature [6]. Sterility in SF females decreases with age as I element repression seems to be established gradually in the hybrids. Fertility is also affected by the application of a heat treatment. When applied to SF females during early oogenesis, fertility decreases; but when applied during late oogenesis, fertility increases. The mechanisms for these two nongenetic factors are still unknown but we know their effects to be cumulative.
Image Submitted by James Tang
Further inquiry into this unique hybrid system could potentially reveal more on the mechanism and rate of I elements and the piRNA systems that regulate their propagation. The results stand to not only tell us more about sterility in these systems, but also regarding the effects of similar transposable elements in other systems as well. For example, one type of retrotransposons densely populate the telomeric regions at the ends of chromosomes, and are responsible for maintaining these regions in fruit flies [7]. Telomeres are essential for cell survival because it caps off the ends of chromosomes and prevents the internal regions that contain valuable information from being lost due to incomplete end replication after each round of mitosis.
In a study of another family of retrotransposons, the LTR family, gypsy elements were observed to increase in concentration in the brains of animals in an age-dependent manner. By disrupting the piRNA repression system, gypsy elements were prematurely unleashed on the brains of these mutants [8]. Aside from finding heightened gypsy levels in the brains of these mutants, early onset memory impairment and shortened lifespans were also observed.
Researchers have estimated that there between 8,000-17,000 retrocopies in the human reference genome that was sequenced back in 2003 [9]. Retrotransposon movement has had and will continue to have profound impacts on our genetic diversity and stability. Understanding how these elements move and the systems that keep them in check is an exciting and an ever-growing field of study.
Authors
References
1. Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300(5617):286-290. doi:10.1126/science.1084564.
2. Human Genome Project FACT SHEET-Human Genome Project.; 2010. http://hapmap.ncbi.nlm.nih.gov/. Accessed August 23, 2018.
3. Jorde LB. Genetic Variation and Human Evolution. https://www.ashg.org/education/pdf/geneticvariation.pdf. Accessed August 23, 2018.
4. Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. doi:10.1242/dev.006486.
5. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. doi:10.1126/science.1165171.
6. Orsi G a, Joyce EF, Couble P, McKim KS, Loppin B. Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. J Cell Sci. 2010;123(Pt 20):3515-3524. doi:10.1242/jcs.073890.
7. Zhang L, Rong YS. Retrotransposons at Drosophila telomeres: host domestication of a selfish element for the maintenance of genome integrity. Biochim Biophys Acta. 2012;1819(7):771-775. doi:10.1016/j.bbagrm.2012.01.018.
8. Li W, Prazak L, Chatterjee N, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16(5):529-531. doi:10.1038/nn.3368.
9. Richardson SR, Salvador-Palomeque C, Faulkner GJ. Diversity through duplication: whole-genome sequencing reveals novel gene retrocopies in the human population. Bioessays. 2014;36(5):475-481. doi:10.1002/bies.201300181.




![Figure 2. IR hybrids exhibit sterility in SF females generated from the dysgenic cross (left), whereas the reciprocal, non-dysgenic cross (right) produces fertile female offspring. Images of flies are by Madboy74 [CC0], retrieved from Wikimedia Comm…](https://images.squarespace-cdn.com/content/v1/5832160cf7e0ab3a3c9604f2/1536965384696-NUAYWB1XU0G0BIRY3RH8/Picture2.png)


